FASTAptamer: A Bioinformatic Toolkit for High-throughput Sequence Analysis of Combinatorial Selections: Molecular Therapy - Nucleic Acids
High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy | Scientific Reports
National Center for Genome Analysis Support: Carrie Ganote Ram Podicheti Le-Shin Wu Tom Doak Quality Control and Assessment. - ppt download
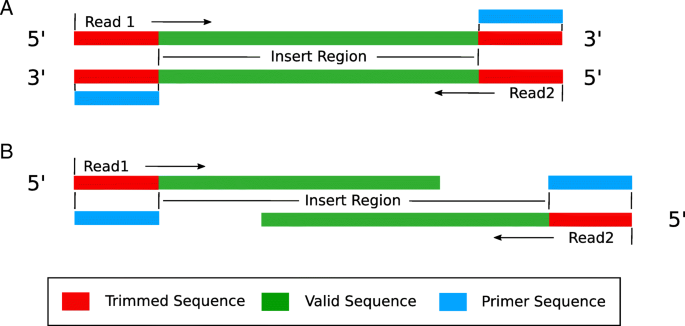
pTrimmer: An efficient tool to trim primers of multiplex deep sequencing data | BMC Bioinformatics | Full Text
From cutadapt to sequencetools (sqt): a versatile toolset for sequencing projects References Relevant Web sites

An open-sourced bioinformatic pipeline for the processing of Next-Generation Sequencing derived nucleotide reads: Identification and authentication of ancient metagenomic DNA | bioRxiv

Handling of targeted amplicon sequencing data focusing on index hopping and demultiplexing using a nested metabarcoding approach in ecology | Scientific Reports
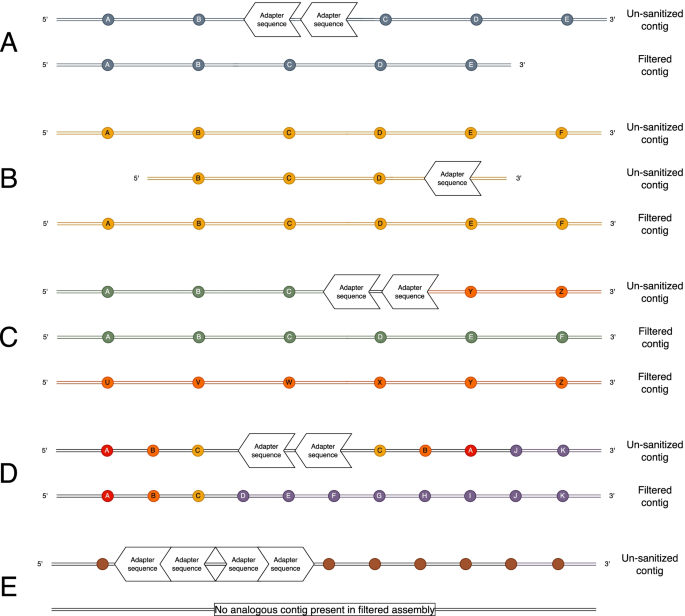


![PDF] Atropos: specific, sensitive, and speedy trimming of sequencing reads | Semantic Scholar PDF] Atropos: specific, sensitive, and speedy trimming of sequencing reads | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d49f5a8963dd71c21ffa29ad631aa51c32af986f/4-Figure1-1.png)